Intelligent DNA Identification System

Hello folks, I know the first thing pop-in your mind What is Intelligent DNA Identification System aka IDIS?
This's the name of my graduation project in 2009, it's just a Bioinformatics system implemented with DNA as biometric system, and used for identification.

Why DNA, Biometrics, and Bioinformatics?
Because we choose our graduation project, we want to do something unique and hard to do.
You said it's a biometric system and used for identification, Are there another applications of biometric systems?
Yep, there're three types of biometric applications:
- Identification: When you want to search for unknown personal, to know any information about him.
- Verification: is to verify XYZ personal is really XYZ personal or not.
- Screening: searching for "XYZ" personal in a list like "Banned from traveling List".
What makes this field Unique and Hard?
The Bioinformatics and Biometrics fields are rare in Egypt, and this was the challenge to make something new and not stable yet, the information for these fields are updating every year maybe less, and the information on the internet are rare and sometimes not free, but we manage to read some IEEE papers and the great book of O'Reilly BLAST.
What is BLAST?
B.L.A.S.T. stand for Basic Local Alignment Search Tool, which our system is based on this powerful tool.

Obstacles
When we decide to build our system, we face some obstacles which are:
- Information availability, it's not easy to find research papers and books about DNA.
- Any implementation written in either Python or Java; and we want to build our system on C#.
- The algorithm of BLAST works for all living being, and we want our system to work only with human being.
So I decide to read the O'Reilly BLAST book from cover to cover, but I really read the first 3 chapters and then an idea popup in my mind, why not customize this algorithm to be optimized for our system.
I used the concept of dynamic programming which is divide the big problem into small problems and solve them, so I created an algorithm that divided into 2 steps:
- Inexact match.
- Exact match.
How does it work?
First let's define the Inputs, Outputs, and Processing.
Input
The unknown personal's DNA
Processing
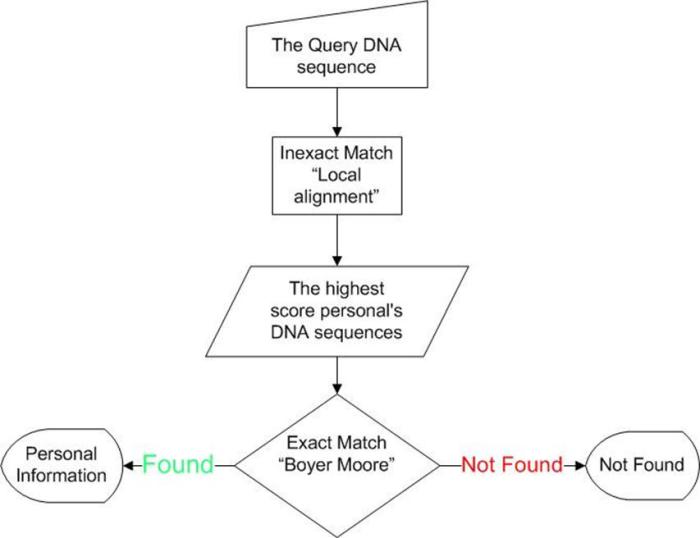
- The first step "Inexact match": take the input DNA and make a local alignment process and select the most 10 persons made the highest match score, who is a one of them is the person, or they are related, so we can use this step to identify the person's relatives like brother, sister, cousin, uncle, aunt, etc.
- The second step "exact match": we use algorithm called 'Boyer-Moore' which used in string search processing, then the inputs for this step are the first step output "10 personal's DNA and the unknown person's DNA, and run process 10 times, every time run the algorithm with the unknown person's DNA and each of the 10 persons, the result either match or mismatch.
Output
- Match "personal's information found".
- Mismatch "personal's information not found".
The Flowchart of the algorithm: